Dedication: For dearest Ruzbeh, and for each and every family out there that has ever felt despair and bewilderment at their (Norrie) child’s neurological/cognitive problems.
Acknowledgements: I thank the following scientists:
- Prof. Yvonne Jones and Dr. Tao-Hsin Chang, Division of Structural Biology, Wellcome Trust Centre for Human Genetics, University of Oxford, for taking the time and trouble to answer all my questions, and for permission to use figure 8B from Chang et al., 2015
- Prof. Jeremy Nathans, Professor of Molecular Biology and Genetics, of Neuroscience and of Ophthalmology, Johns Hopkins University School of Medicine, for providing clarifications to my queries related to Smallwood et al., 2007
- Dr. Jiyuan Ke, Research Scientist, Van Andel Research Institute, for permission to use figures 2A, 2B, and 2C from Ke et al., 2013
I would also like to thank families who have provided me with data pertaining to their (Norrie) child.
Here’s the thing – in Norrie Disease (ND) related literature, the number of cases presenting brain related problems (cognitive impairment, autistic traits, psychosis) is listed as up to 30-50%. This is a not a small percentage. Yet every once in a while, I get asked - How do you know your son’s mental problems are related to Norrie Disease? It might be something else.
An image of Ruzbeh – this amazingly intelligent person who, equally amazingly, is …….what? …….cognitively impaired? ...........intellectually impaired? …......has sensory integration problems? ………..I don’t even know what name I should give to it! – appears in my mind and I find myself saying - I don’t know that they are (related), I just assumed that they are.
Well, my assumption is hardly scientific proof, is it?
And so it was that I decided to take out some time to study Ruzbeh’s genetic mutation. Ruzbeh’s mutation is a missense mutation, which is the simplest type of mutation possible. A missense mutation implies that one amino acid (just one) of the protein has got substituted by another amino acid.
I requested members of the online Norrie support group for additional missense ND mutations, so that I could analyze all the mutations together for comparison.
I read through three research articles which, directly or indirectly, deal with missense mutations of Norrin. These are: Smallwood et al, 2007, Ke et al., 2013, and Chang et al., 2015.
I was amazed at what I learnt.
Before I get to my findings though, I thought you might like to read the following:
- a recap of some basic facts about genes, proteins, and amino acids
- definition of a missense mutation
- the composition, structure, and functioning of the protein Norrin
An organism's genetic blueprint is encoded by the molecule called Deoxyribonucleic acid (DNA). DNA is a linear molecule composed of four types of smaller chemical molecules called nucleotide bases: adenine (A), cytosine (C), guanine (G), and thymine (T). The order of these bases is called the DNA sequence.
Segments of DNA that carry genetic information are called genes, and they are inherited by offspring from their parents during reproduction. Each gene is a coded description for making a particular protein. The sequence of bases in the gene determines the protein that it codes for.
Proteins perform a large chunk of the important jobs a cell needs to do. They make up our muscles, hair, nails; they make chemical reactions happen; they digest our food. Protein molecules are long chains of amino acids that are folded into a three-dimensional shape. The shape a protein takes is incredibly important for its function. Think about it - if a key has the wrong shape, it can't open a lock, right? The same is true for a protein and its function.
Amino acids are small molecules that are the building blocks of proteins. There are 20 amino acids & they have names like leucine, methionine, tryptophan, etc. Each amino acid is denoted either by a 3 letter abbreviation or by a 1 letter code. So leucine is Leu or L, methionine is Met or M, and tryptophan is Trp or W.
The genetic code is a set of three-letter combinations (called codons) of the four nucleotide bases (A, C, T, G), each of which corresponds to a specific amino acid or stop signal.
There are 64 possible permutations, or combinations, of three-letter sequences that can be made from the four nucleotides. Of these 64 codons, 61 represent amino acids, and three are stop signals. Stop codons serve as a signal that the end of the chain has been reached during protein synthesis.
Given below is a table depicting the genetic code.
The colors highlight the fact that most amino acids have more than one code. For example, Serine (Ser) has 6 codes.
The 1 letter code corresponding to each amino acid is as shown below:
| Alanine | Ala | A |
| Arginine | Arg | R |
| Asparagine | Asn | N |
| Aspartic Acid | Asp | D |
| Cysteine | Cys | C |
| Glutamine | Gln | Q |
| Glutamic Acid | Glu | E |
| Glycine | Gly | G |
| Histidine | His | H |
| Isoleucine | Ile | I |
| Leucine | Leu | L |
| Lysine | Lys | K |
| Methionine | Met | M |
| Phenylalanine | Phe | F |
| Proline | Pro | P |
| Serine | Ser | S |
| Threonine | Thr | T |
| Tryptophan | Trp | W |
| Tyrosine | Tyr | Y |
| Valine | Val | V |
When amino acids combine to form a protein, they are also referred to as amino acid residues, or simply residues.
As an example, consider the peptide (a peptide is simply a short chain of amino acids) Gly-Ser-Gln-Met-Val-Ile or GSQMVI. The 1st residue (G1) is the amino acid glycine (Gly), the 5th residue (V5) is the amino acid valine (Val), and so on.
Based on the propensity of an amino acid to be in contact with water, it may be classified as hydrophobic (low propensity to be in contact with water), neutral, or hydrophilic.
(2) Missense mutations [Source: Wikipedia]
A mutation is a change in a genetic sequence. Mutations include changes as small as the substitution of a single DNA building block, or nucleotide base, with another nucleotide base. Along with substitutions, mutations can also be caused by insertions, deletions, or duplications of DNA sequences. Mutations can be introduced due to mistakes made during DNA replication or due to exposure to mutagens, which are chemical and environmental agents that can introduce mutations in the DNA sequence.
A missense mutation is a genetic change which results in one amino acid of a protein getting substituted by another amino acid.
For example, a change in just one nucleotide base can result in a missense mutation, as shown below:
(3) A brief note on the composition, structure & function of Norrin
The Norrie Disease gene (Ndp) codes for the protein Norrin.
Norrin comprises of 133 residues. These are listed below:
KTDSSFIMDSDPRRCMRHHYVDSISHPLYKCSSKMVLLARCEGHCSQASRSEPLVSFSTVLKQPFRSSCHCCRPQTSKLKALRLRCSGGMRLTATYRYILSCHCEECNS
Norrin structure
The crystal structure of human Norrin reveals a dimer formation. A dimer consists of two structurally similar molecules, each of which is referred to as a monomer. Norrin is a homodimer, which means that the two monomers are identical. The following is a ribbon diagram of the Norrin dimer consisting of two identical monomers in green and magenta:
[Source: Ke et al., 2013 Figure 2B]
Each Norrin monomer contains 11 cysteines (Cys or C) – C39, C55, C65, C69, C93, C95, C96, C110, C126, C128 & C131. These are shown in red below:
KTDSSFIMDSDPRRCMRHHYVDSISHPLYKCSSKMVLLARCEGHCSQASRSEPLVSFSTVLKQPFRSSCHCCRPQTSKLKALRLRCSGGMRLTATYRYILSCHCEECNS
Cysteine has a sulfur atom (the chemical formula for cysteine being C3H7NO2S). Sulfur atoms on two separate cysteine residues can bond with each other, forming a disulfide bridge or a disulphide bond.
Schematic representation of disulphide bonds in a protein [Source: Wikipedia]
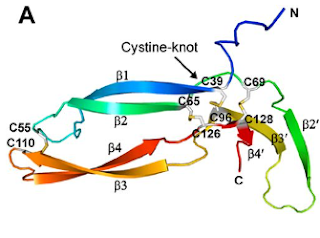
Of the eleven cysteines of the Norrin monomer, six cysteines form three pairs of intramolecular (i.e. contained within the Norrin monomer) disulphide bonds (C39–C96, C65–C126, C69–C128) which result in the protein folding into a cystine knot structure.
Diagram depicting the cystine knot motif of Norrin. A: Amino acid sequence of norrin. Highlighted C's indicate cysteine residues involved in disulfide bonds which lead to the cystine knot structure. B: Structure of Norrin. The roman numerals label the cysteine residues involved in cystine knot disulfide bonds (connecting lines).
[Source: K Drenser et al., 2007]
Norrin function depends critically on the three pairs of cysteines that form the highly conserved trio of disulfide bonds shared among all cystine knot proteins (Smallwood et al, 2007).
Another pair of cysteines (C55–C110) forms a fourth intramolecular disulphide bond.
The following diagram shows all four intramolecular disulfide bonds (C39–C96, C65–C126, C69–C128, and C55–C110) in the Norrin monomer:
[Source: Ke et al., 2013 Figure 2A]
Additionally, there exist three intermolecular (i.e. between the two monomers of the Norrin dimer) disulphide bonds (C93–C95, C95–C93, and C131–C131). These disulphide bonds are important for the stability of the Norrin dimer.
The following is a diagram of the Norrin dimer, showing the three intermolecular disulfide bonds (C93–C95, C95–C93, and C131–C131):
[Source: Ke et al., 2013 Figure 2C]
In addtition to these three cysteine residues, the Norrin dimer interface comprises of numerous other residues. The dimeric interface of Norrin is important for its function.
Functioning of Norrin
The cells within an animal body need to be able to communicate with each other to coordinate many complex processes in the body, such as the formation of tissues and organs. One way in which the cells can communicate is through a pathway called Wnt signaling.
Norrin activates the Wnt signaling pathway by binding (attaching) to two other proteins/molecules - Frizzled-4 (also written as Fzd-4 or Fzd4) and Lrp5/6. For the purpose of this blog, we don’t really need to know a great deal about these molecules. Suffice it to say that Fzd4 and Lrp5/6 are proteins coded for by the genes FZD4, LRP5, and LRP6, in much the same way that Norrin is a protein coded for by the gene NDP. In scientific parlance, Fzd4 and Lrp5/6 are referred to as receptors of Norrin.
Norrin has distinct binding sites for Fzd4 and for Lrp5/6. The binding site for Fzd4 and a potential binding site for Lrp5/6 have been mapped out in Chang et al., 2015, as shown in the diagram below. The two concave shapes in the diagram represent the two monomers of Norrin.
Diagram depicting the various binding sites on Norrin. The two concave shapes represent the two monomers of Norrin. GAG (glycosaminoglycan) is yet another type of molecule which binds to the Norrin-Fzd4 complex. [Source: Chang et al., 2015]
The following table categorizes residues of Norrin based on the structure and/or function of the protein:
| Category | Residues |
| Cystine knot motif | C39, C65, C69, C96, C126, C128 |
| Fzd-4 binding site | D33, S44, P36, R38, residues 40-43, V45, D46, S47, M59, V60, L61, T100, K102, K104, A105, L106, Y120, Y122, L124, S125 |
| Lrp5/6 binding site | K54, R90, R97, G112, R121 |
| GAG binding site | K58, R107, R109, R115 |
| Dimer interface | Y44, I48, S49, H50, P51, residues 62-81, residues 83-85, P88, F89, S91, residues 93-96, P98, S101, L103, L108, residues 116-119, I123, C131, N132 |
| Other | Any residue of Norrin which is not listed in any of the above five categories |
(4) Findings
I had four mutations to analyze:
- C69Y (the 69th residue has got substituted by tyrosine in place of cysteine)
- R90C (the 90th residue has got substituted by cysteine in place of arginine)
- F81L (the 81st residue has got substituted by leucine in place of phenylalanine)
- G112Q (the 112th residue has got substituted by glutamine in place of glycine)
C69Y: This is Ruzbeh’s mutation. C69!! This is one of the six cysteines which form the disulphide bonds leading to the cystine knot motif of Norrin. I presume this mutation would dramatically alter the Norrin signaling activity.
R90C, F81L, G112Q: These mutations were provided by families from the Norrie support group. These are easily identified as mutations on the (potential) Lrp5/6 binding site (residues R90 and G112) and on the dimer interface (residue F81).
| Missense mutation | Category of residue* | Age of person / Symptoms seen** |
| C69Y | One of the six cysteines which form the cystine knot motif of Norrin. | 18 years (Ruzbeh).
Congenital blindness, hearing loss, autistic behavior, cognitive impairment, few psychotic episodes, two seizures. |
| R90C | Lrp5/6 binding site | 8 years.
Congenital blindness, few language difficulties, sensory sensitive. |
| F81L | Dimer interface | 4 years.
Congenital blindness, developmental delay, mild learning difficulties. |
| G112Q | Lrp5/6 binding site | 22 years.
Blind in one eye due to retina detachment at age 2 ½ years old. 20/30 vision in other eye post vitrectomy surgery at age 4. |
* Source: Chang et al., 2015
** Data provided by families
So far so good.
I wanted to go a step further and find some quantitative data for these mutations. In separate communications from Prof. Nathans and from Prof. Jones, I have come to know that quantitative data for Norrin’s signaling activity for different mutations, does not exist. However, they did have this to say:
C69Y (Ruzbeh’s mutation):
Prof. Nathans agrees that C69 being a critical residue of Norrin, this mutation would result in a severe loss of function of the protein Norrin.
Prof. Jones: Not all residues in a protein have equally important roles. Some missense mutations are likely to have more effect because of the position of the residue in the 3D structure of the protein and/or because of the extent of the change the missense mutation makes to the properties of the residue. For example, a particular residue can be essential for a protein to fold into its correct 3D shape, and/or to maintain that shape. This category of residue is important because if the protein does not have the correct overall shape it cannot work properly. The six cysteine residues which form the cystine-knot motif of Norrin do so because, uniquely, one cysteine can bond to another cysteine to form a disulphide bond, and the three disulphide bonds of the cystine-knot motif are what locks one copy of Norrin into its correct 3D shape.
Sadly, the extent of your son’s symptoms is consistent with this being a missense mutation affecting the cystine-knot motif and so stopping the protein forming the correct 3D shape and therefore stopping its ability to bind to both its receptors.
R90C, F81L, G112Q
Prof. Jones: Norrin works as a dimer so the residues that contribute to the dimer interface between the two copies of Norrin are important.
Three cysteine residues per copy of Norrin hold two copies together by forming disulphide bonds at the dimer interface. Because of the unique ability of cysteine to form a disulphide bond, and because of the role of disulphide bridges in stabilising the correct 3D structure of Norrin, missense mutations of these residues are likely to be serious.
How serious missense mutations are of other residues at the dimer interface, or of the residues that Norrin uses to bind to its receptors Fzd4 and Lrp5/6, depends on whether the missense mutation makes a big change to essential properties of the residue.
R90C and G112Q both result in big changes to the properties of the affected residues but we do not know how these particular properties contribute to the binding of Norrin to Lrp5/6 because we do not have the structure of Norrin bound to Lrp5/6. Also, without a structure, we do not know whether the residue is at the centre of the binding site, often a residue at the centre of the binding site will make more of a contribution than one at the edge. The only thing we can say now is that we do not expect these residues to affect the overall 3D shape of the protein or its ability to bind to Lrp5/6.
If we take a residue that contributes to the ability of Norrin to bind to one of its receptors, or to
form the dimer interface, because it is a ‘hydrophobic’ residue (i.e. ‘sticky’) then a missense mutation that changes the residue to one that is slightly different in size, but still hydrophobic/sticky, might not have such a serious effect – the dimer may be slightly distorted or the binding to the receptor may be weaker, so the signaling activity is reduced, but it is not lost completely.
F81L is at the dimer interface of Norrin. F (phenylalanine) is a sticky/hydrophobic residue, and so is L (leucine), albeit F is a bit bigger than L. We think that the dimer is likely to still form but it may be a little distorted so the binding of F81L Norrin to its receptors may be affected because it is not quite the same 3D shape as usual.
So there you have it. In black and white.
And I finally know. The reason why.
Till next time then,
Meenu.